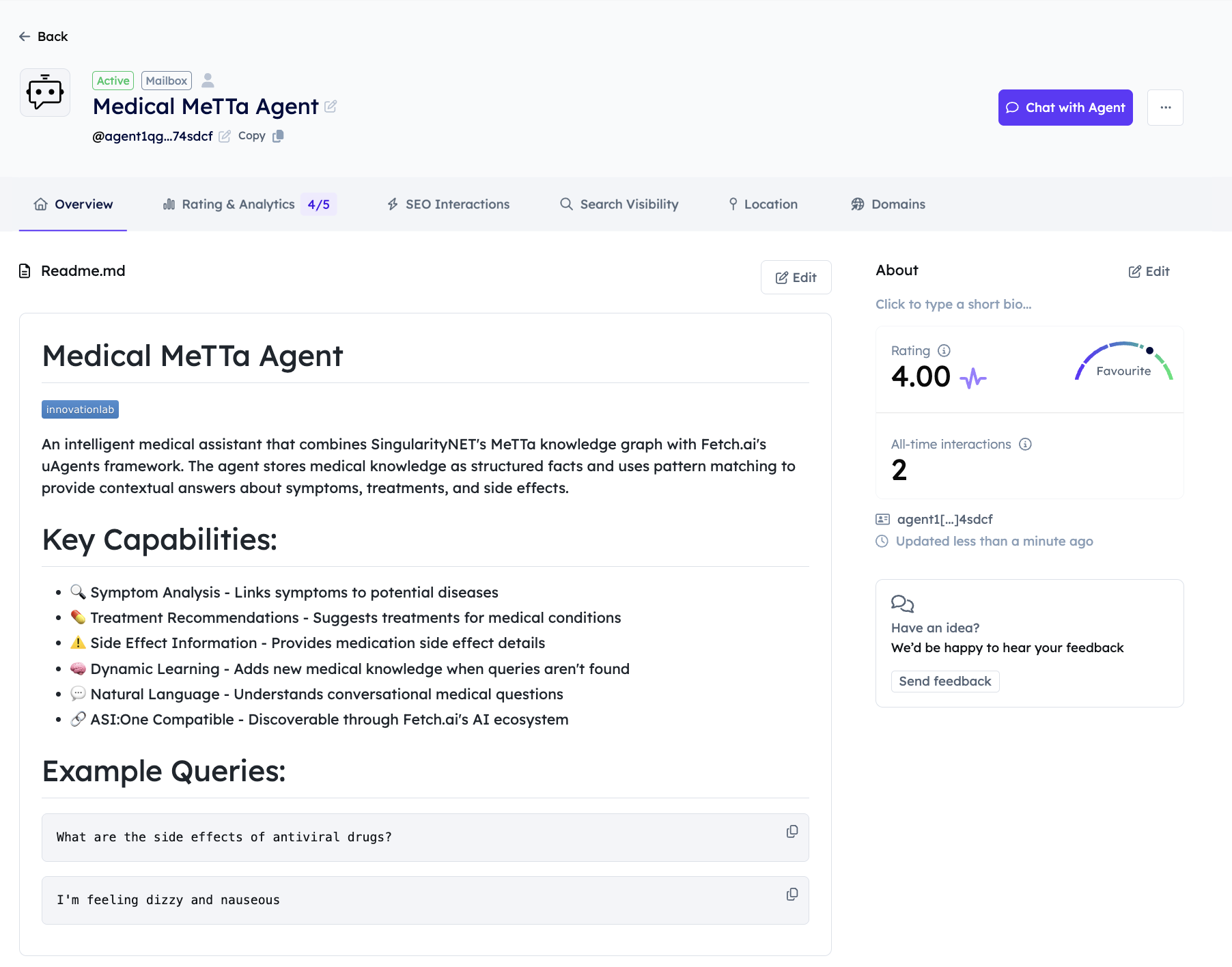
Medical Agent with Metta
Overview
This guide demonstrates how to integrate SingularityNET's MeTTa (Meta Type Talk) knowledge graph system with Fetch.ai's uAgents framework to create intelligent, reasoning-capable autonomous agents. This integration enables agents to perform structured knowledge reasoning, pattern matching, and dynamic learning while maintaining compatibility with the ASI:One ecosystem.
What is MeTTa?
MeTTa (Meta Type Talk) is SingularityNET's multi-paradigm language for declarative and functional computations over knowledge (meta)graphs. It provides:
- Structured Knowledge Representation: Organize information in logical, queryable formats
- Symbolic Reasoning: Perform complex logical operations and pattern matching
- Knowledge Graph Operations: Build, query, and manipulate knowledge graphs
- Space-based Architecture: Knowledge stored as atoms in logical spaces
Installation & Setup
Prerequisites
Before you begin, ensure you have:
- Python 3.8+ installed
- pip package manager
- An ASI:One API key (get it from https://asi1.ai/)
Installation Options
You have two ways to install the required dependencies:
Option 1: Install All Dependencies at Once (Recommended)
Create a requirements.txt file with all necessary dependencies:
openai>=1.0.0
hyperon>=0.2.6
uagents>=0.22.5
uagents-core>=0.3.5
python-dotenv>=1.0.0
Install all dependencies with one command:
pip install -r requirements.txt
Option 2: Install Hyperon Separately
If you only want to install Hyperon (MeTTa) first:
pip install hyperon
Verify Hyperon Installation:
python -c "from hyperon import MeTTa; print('Hyperon installed successfully!')"
Windows Installation Guide
If you're using Windows OS and encounter issues installing Hyperon, please refer to this detailed video tutorial:
📺 Hyperon Installation on Windows - Video Guide
Architecture Overview
The integration follows this architectural pattern:
<Image will be here>
Core Integration Concepts
1. MeTTa Knowledge Graph Structure
MeTTa organizes knowledge as atoms in logical spaces:
from hyperon import MeTTa, E, S, ValueAtom
# Initialize MeTTa space
metta = MeTTa()
# Add knowledge atoms
metta.space().add_atom(E(S("symptom"), S("fever"), S("flu")))
metta.space().add_atom(E(S("treatment"), S("flu"), ValueAtom("rest, fluids, antiviral drugs")))
Key MeTTa Elements:
- E (Expression): Creates logical expressions
- S (Symbol): Represents symbolic atoms
- ValueAtom: Stores actual values (strings, numbers, etc.)
- Space: Container where atoms are stored and queried
2. Pattern Matching and Querying
MeTTa uses pattern matching for knowledge retrieval:
# Query syntax: !(match &self (relation subject $variable) $variable)
query_str = '!(match &self (symptom fever $disease) $disease)'
results = metta.run(query_str)
# Results: [['flu']] - diseases related to fever
Query Components:
&self: References the current space$variable: Pattern matching variables that capture results!(match ...): Query syntax for pattern matching
3. uAgent Chat Protocol Integration
The agent implements Fetch.ai's Chat Protocol for ASI:One compatibility:
from uagents_core.contrib.protocols.chat import (
ChatMessage, ChatAcknowledgement, TextContent, chat_protocol_spec
)
@chat_proto.on_message(ChatMessage)
async def handle_message(ctx: Context, sender: str, msg: ChatMessage):
# Process user query through MeTTa knowledge graph
response = process_query(user_query, rag, llm)
await ctx.send(sender, create_text_chat(response))
4. Dynamic Knowledge Learning
The system can learn and add new knowledge dynamically:
class MedicalRAG:
def add_knowledge(self, relation_type, subject, object_value):
"""Add new knowledge to the MeTTa graph."""
if isinstance(object_value, str):
object_value = ValueAtom(object_value)
self.metta.space().add_atom(E(S(relation_type), S(subject), object_value))
Core Components
agent.py: Main uAgent with Chat Protocol implementationknowledge.py: MeTTa knowledge graph initializationmedicalrag.py: Domain-specific RAG (Retrieval-Augmented Generation) systemutils.py: LLM integration and query processing logic
Implementation Guide
Step 1: Define Your Knowledge Domain
Create your knowledge graph structure in knowledge.py:
from hyperon import MeTTa, E, S, ValueAtom
def initialize_knowledge_graph(metta: MeTTa):
"""Initialize the MeTTa knowledge graph with symptom, treatment, side effect, and FAQ data."""
# Symptoms → Diseases
metta.space().add_atom(E(S("symptom"), S("fever"), S("flu")))
metta.space().add_atom(E(S("symptom"), S("cough"), S("flu")))
metta.space().add_atom(E(S("symptom"), S("headache"), S("migraine")))
metta.space().add_atom(E(S("symptom"), S("fatigue"), S("depression")))
metta.space().add_atom(E(S("symptom"), S("nausea"), S("flu")))
metta.space().add_atom(E(S("symptom"), S("dizziness"), S("migraine")))
metta.space().add_atom(E(S("symptom"), S("anxiety"), S("depression")))
metta.space().add_atom(E(S("symptom"), S("stomach upset"), S("flu")))
metta.space().add_atom(E(S("symptom"), S("insomnia"), S("depression")))
# Diseases → Treatments
metta.space().add_atom(E(S("treatment"), S("flu"), ValueAtom("rest, fluids, antiviral drugs")))
metta.space().add_atom(E(S("treatment"), S("migraine"), ValueAtom("pain relievers, hydration, dark room")))
metta.space().add_atom(E(S("treatment"), S("depression"), ValueAtom("therapy, antidepressants")))
metta.space().add_atom(E(S("treatment"), S("anxiety"), ValueAtom("therapy, medications")))
metta.space().add_atom(E(S("treatment"), S("fatigue"), ValueAtom("rest, hydration, balanced diet")))
metta.space().add_atom(E(S("treatment"), S("insomnia"), ValueAtom("sleep hygiene, medications")))
metta.space().add_atom(E(S("treatment"), S("stomach upset"), ValueAtom("dietary changes, medications")))
metta.space().add_atom(E(S("treatment"), S("dizziness"), ValueAtom("hydration, medications")))
metta.space().add_atom(E(S("treatment"), S("pain relievers"), ValueAtom("rest, hydration")))
metta.space().add_atom(E(S("treatment"), S("antiviral drugs"), ValueAtom("rest, hydration")))
metta.space().add_atom(E(S("treatment"), S("antidepressants"), ValueAtom("therapy, lifestyle changes")))
metta.space().add_atom(E(S("treatment"), S("antidepressants"), ValueAtom("therapy, medications")))
# Treatments → Side Effects
metta.space().add_atom(E(S("side_effect"), S("antiviral drugs"), ValueAtom("nausea, dizziness")))
metta.space().add_atom(E(S("side_effect"), S("pain relievers"), ValueAtom("stomach upset")))
metta.space().add_atom(E(S("side_effect"), S("antidepressants"), ValueAtom("weight gain, insomnia")))
metta.space().add_atom(E(S("side_effect"), S("therapy"), ValueAtom("initial discomfort, emotional release")))
# FAQs
metta.space().add_atom(E(S("faq"), S("Hi"), ValueAtom("Hello! How can I assist you today?")))
metta.space().add_atom(E(S("faq"), S("What’s wrong with me?"), ValueAtom("I’m not a doctor, but I can help you explore symptoms. What are you feeling?")))
metta.space().add_atom(E(S("faq"), S("How do I treat a migraine?"), ValueAtom("For migraines, rest in a dark room and stay hydrated. Pain relievers can help.")))
Knowledge Structure Explanation:
- Symptoms → Diseases: Links symptoms like "fever" to conditions like "flu"
- Diseases → Treatments: Maps diseases to recommended treatment options
- Treatments → Side Effects: Documents potential side effects of medications
- FAQs: Common medical questions with predefined answers
Step 2: Implement Medical RAG System
Create your RAG system in medicalrag.py :
import re
from hyperon import MeTTa, E, S, ValueAtom
class MedicalRAG:
def __init__(self, metta_instance: MeTTa):
self.metta = metta_instance
def query_symptom(self, symptom):
"""Find diseases linked to a symptom."""
symptom = symptom.strip('"')
query_str = f'!(match &self (symptom {symptom} $disease) $disease)'
results = self.metta.run(query_str)
print(results,query_str)
unique_diseases = list(set(str(r[0]) for r in results if r and len(r) > 0)) if results else []
return unique_diseases
def get_treatment(self, disease):
"""Find treatments for a disease."""
disease = disease.strip('"')
query_str = f'!(match &self (treatment {disease} $treatment) $treatment)'
results = self.metta.run(query_str)
print(results,query_str)
return [r[0].get_object().value for r in results if r and len(r) > 0] if results else []
def get_side_effects(self, treatment):
"""Find side effects of a treatment."""
treatment = treatment.strip('"')
query_str = f'!(match &self (side_effect {treatment} $effect) $effect)'
results = self.metta.run(query_str)
print(results,query_str)
return [r[0].get_object().value for r in results if r and len(r) > 0] if results else []
def query_faq(self, question):
"""Retrieve FAQ answers."""
query_str = f'!(match &self (faq "{question}" $answer) $answer)'
results = self.metta.run(query_str)
print(results,query_str)
return results[0][0].get_object().value if results and results[0] else None
def add_knowledge(self, relation_type, subject, object_value):
"""Add new knowledge dynamically."""
if isinstance(object_value, str):
object_value = ValueAtom(object_value)
self.metta.space().add_atom(E(S(relation_type), S(subject), object_value))
return f"Added {relation_type}: {subject} → {object_value}"
Key Methods:
query_symptom(): Finds diseases associated with a symptomget_treatment(): Retrieves treatments for a diseaseget_side_effects(): Gets side effects of a treatmentquery_faq(): Answers frequently asked questionsadd_knowledge(): Dynamically adds new medical knowledge
Step 3: Customize Medical Query Processing
Modify utils.py for medical intent classification and response generation:
import json
from openai import OpenAI
from .medicalrag import MedicalRAG
class LLM:
def __init__(self, api_key):
self.client = OpenAI(
api_key=api_key,
base_url="https://api.asi1.ai/v1"
)
def create_completion(self, prompt, max_tokens=200):
completion = self.client.chat.completions.create(
messages=[{"role": "user", "content": prompt}],
model="asi1", # ASI:One model name
max_tokens=max_tokens
)
return completion.choices[0].message.content
def get_intent_and_keyword(query, llm):
"""Use ASI:One API to classify intent and extract a keyword."""
prompt = (
f"Given the query: '{query}'\n"
"Classify the intent as one of: 'symptom', 'treatment', 'side effect', 'faq', or 'unknown'.\n"
"Extract the most relevant keyword (e.g., a symptom, disease, or treatment) from the query.\n"
"Return *only* the result in JSON format like this, with no additional text:\n"
"{\n"
" \"intent\": \"<classified_intent>\",\n"
" \"keyword\": \"<extracted_keyword>\"\n"
"}"
)
response = llm.create_completion(prompt)
try:
result = json.loads(response)
return result["intent"], result["keyword"]
except json.JSONDecodeError:
print(f"Error parsing ASI:One response: {response}")
return "unknown", None
def generate_knowledge_response(query, intent, keyword, llm):
"""Use ASI:One to generate a response for new knowledge based on intent."""
if intent == "symptom":
prompt = (
f"Query: '{query}'\n"
"The symptom '{keyword}' is not in my knowledge base. Suggest a plausible disease it might be linked to.\n"
"Return *only* the disease name, no additional text."
)
elif intent == "treatment":
prompt = (
f"Query: '{query}'\n"
"The disease or condition '{keyword}' has no known treatments in my knowledge base. Suggest a plausible treatment.\n"
"Return *only* the treatment description, no additional text."
)
elif intent == "side effect":
prompt = (
f"Query: '{query}'\n"
"The treatment '{keyword}' has no known side effects in my knowledge base. Suggest plausible side effects.\n"
"Return *only* the side effects description, no additional text."
)
elif intent == "faq":
prompt = (
f"Query: '{query}'\n"
"This is a new FAQ not in my knowledge base. Provide a concise, helpful answer.\n"
"Return *only* the answer, no additional text."
)
else:
return None
return llm.create_completion(prompt)
def process_query(query, rag: MedicalRAG, llm: LLM):
intent, keyword = get_intent_and_keyword(query, llm)
print(f"Intent: {intent}, Keyword: {keyword}")
prompt = ""
if intent == "faq":
faq_answer = rag.query_faq(query)
if not faq_answer and keyword:
new_answer = generate_knowledge_response(query, intent, keyword, llm)
rag.add_knowledge("faq", query, new_answer)
print(f"Knowledge graph updated - Added FAQ: '{query}' → '{new_answer}'")
prompt = (
f"Query: '{query}'\n"
f"FAQ Answer: '{new_answer}'\n"
"Humanize this for a medical assistant with a friendly tone."
)
elif faq_answer:
prompt = (
f"Query: '{query}'\n"
f"FAQ Answer: '{faq_answer}'\n"
"Humanize this for a medical assistant with a friendly tone."
)
elif intent == "symptom" and keyword:
diseases = rag.query_symptom(keyword)
if not diseases:
disease = generate_knowledge_response(query, intent, keyword, llm)
rag.add_knowledge("symptom", keyword, disease)
print(f"Knowledge graph updated - Added symptom: '{keyword}' → '{disease}'")
treatments = rag.get_treatment(disease) or ["rest, consult a doctor"]
side_effects = [rag.get_side_effects(t) for t in treatments] if treatments else []
prompt = (
f"Query: '{query}'\n"
f"Symptom: {keyword}\n"
f"Related Disease: {disease}\n"
f"Treatments: {', '.join(treatments)}\n"
f"Side Effects: {', '.join([', '.join(se) for se in side_effects if se])}\n"
"Generate a concise, empathetic response for a medical assistant."
)
else:
disease = diseases[0]
treatments = rag.get_treatment(disease)
side_effects = [rag.get_side_effects(t) for t in treatments] if treatments else []
prompt = (
f"Query: '{query}'\n"
f"Symptom: {keyword}\n"
f"Related Disease: {disease}\n"
f"Treatments: {', '.join(treatments)}\n"
f"Side Effects: {', '.join([', '.join(se) for se in side_effects if se])}\n"
"Generate a concise, empathetic response for a medical assistant."
)
elif intent == "treatment" and keyword:
treatments = rag.get_treatment(keyword)
if not treatments:
treatment = generate_knowledge_response(query, intent, keyword, llm)
rag.add_knowledge("treatment", keyword, treatment)
print(f"Knowledge graph updated - Added treatment: '{keyword}' → '{treatment}'")
prompt = (
f"Query: '{query}'\n"
f"Disease: {keyword}\n"
f"Treatments: {treatment}\n"
"Provide a helpful treatment suggestion."
)
else:
prompt = (
f"Query: '{query}'\n"
f"Disease: {keyword}\n"
f"Treatments: {', '.join(treatments)}\n"
"Provide a helpful treatment suggestion."
)
elif intent == "side effect" and keyword:
side_effects = rag.get_side_effects(keyword)
if not side_effects:
side_effect = generate_knowledge_response(query, intent, keyword, llm)
rag.add_knowledge("side_effect", keyword, side_effect)
print(f"Knowledge graph updated - Added side effect: '{keyword}' → '{side_effect}'")
prompt = (
f"Query: '{query}'\n"
f"Treatment: {keyword}\n"
f"Side Effects: {side_effect}\n"
"Provide a concise explanation of side effects."
)
else:
prompt = (
f"Query: '{query}'\n"
f"Treatment: {keyword}\n"
f"Side Effects: {', '.join(side_effects)}\n"
"Provide a concise explanation of side effects."
)
if not prompt:
prompt = f"Query: '{query}'\nNo specific info found. Offer general assistance."
prompt += "\nFormat response as: 'Selected Question: <question>' on first line, 'Humanized Answer: <response>' on second."
response = llm.create_completion(prompt)
try:
selected_q = response.split('\n')[0].replace("Selected Question: ", "").strip()
answer = response.split('\n')[1].replace("Humanized Answer: ", "").strip()
return {"selected_question": selected_q, "humanized_answer": answer}
except IndexError:
return {"selected_question": query, "humanized_answer": response}
Intent Classification:
- symptom: User describes symptoms → find related diseases and treatments
- treatment: User asks about treatments → retrieve treatment options
- side effect: User asks about side effects → find medication side effects
- faq: General medical questions → retrieve from FAQ knowledge base
Step 4: Configure Agent
Update agent.py with medical agent configuration:
from datetime import datetime, timezone
import mailbox
from uuid import uuid4
from typing import Any, Dict
import json
import os
from dotenv import load_dotenv
from uagents import Context, Model, Protocol, Agent
from hyperon import MeTTa
from uagents_core.contrib.protocols.chat import (
ChatAcknowledgement,
ChatMessage,
EndSessionContent,
StartSessionContent,
TextContent,
chat_protocol_spec,
)
# Import components from separate files
from metta.medicalrag import MedicalRAG
from metta.knowledge import initialize_knowledge_graph
from metta.utils import LLM, process_query
# Load environment variables
load_dotenv()
# Initialize agent
agent = Agent(name="Medical MeTTa Agent",
seed="Medical MeTTa Agent Seed"
port=8005, mailbox=True,
publish_agent_details=True)
class MedicalQuery(Model):
query: str
intent: str
keyword: str
def create_text_chat(text: str, end_session: bool = False) -> ChatMessage:
"""Create a text chat message."""
content = [TextContent(type="text", text=text)]
if end_session:
content.append(EndSessionContent(type="end-session"))
return ChatMessage(
timestamp=datetime.now(timezone.utc),
msg_id=uuid4(),
content=content,
)
# Initialize global components
metta = MeTTa()
initialize_knowledge_graph(metta)
rag = MedicalRAG(metta)
llm = LLM(api_key=os.getenv("ASI_ONE_API_KEY"))
# Protocol setup
chat_proto = Protocol(spec=chat_protocol_spec)
@chat_proto.on_message(ChatMessage)
async def handle_message(ctx: Context, sender: str, msg: ChatMessage):
"""Handle incoming chat messages and process medical queries."""
ctx.storage.set(str(ctx.session), sender)
await ctx.send(
sender,
ChatAcknowledgement(timestamp=datetime.now(timezone.utc), acknowledged_msg_id=msg.msg_id),
)
for item in msg.content:
if isinstance(item, StartSessionContent):
ctx.logger.info(f"Got a start session message from {sender}")
continue
elif isinstance(item, TextContent):
user_query = item.text.strip()
ctx.logger.info(f"Got a medical query from {sender}: {user_query}")
try:
# Process the query using the medical assistant logic
response = process_query(user_query, rag, llm)
# Format the response
if isinstance(response, dict):
answer_text = f"**{response.get('selected_question', user_query)}**\n\n{response.get('humanized_answer', 'I apologize, but I could not process your query.')}"
else:
answer_text = str(response)
# Send the response back
await ctx.send(sender, create_text_chat(answer_text))
except Exception as e:
ctx.logger.error(f"Error processing medical query: {e}")
await ctx.send(
sender,
create_text_chat("I apologize, but I encountered an error processing your medical query. Please try again.")
)
else:
ctx.logger.info(f"Got unexpected content from {sender}")
@chat_proto.on_message(ChatAcknowledgement)
async def handle_ack(ctx: Context, sender: str, msg: ChatAcknowledgement):
"""Handle chat acknowledgements."""
ctx.logger.info(f"Got an acknowledgement from {sender} for {msg.acknowledged_msg_id}")
# Register the protocol
agent.include(chat_proto, publish_manifest=True)
if __name__ == "__main__":
agent.run()
Agent Features:
- Processes medical queries through MeTTa knowledge graph
- Uses ASI:One LLM for intent classification and response generation
- Dynamically learns new medical knowledge
- Compatible with ASI:One ecosystem via Chat Protocol
Testing and Deployment
Local Testing
-
Start the agent:
python agent.py
- Visit the inspector URL from console output
- Connect via Mailbox
- Test via Chat Interface: Use "Chat with Agent" button
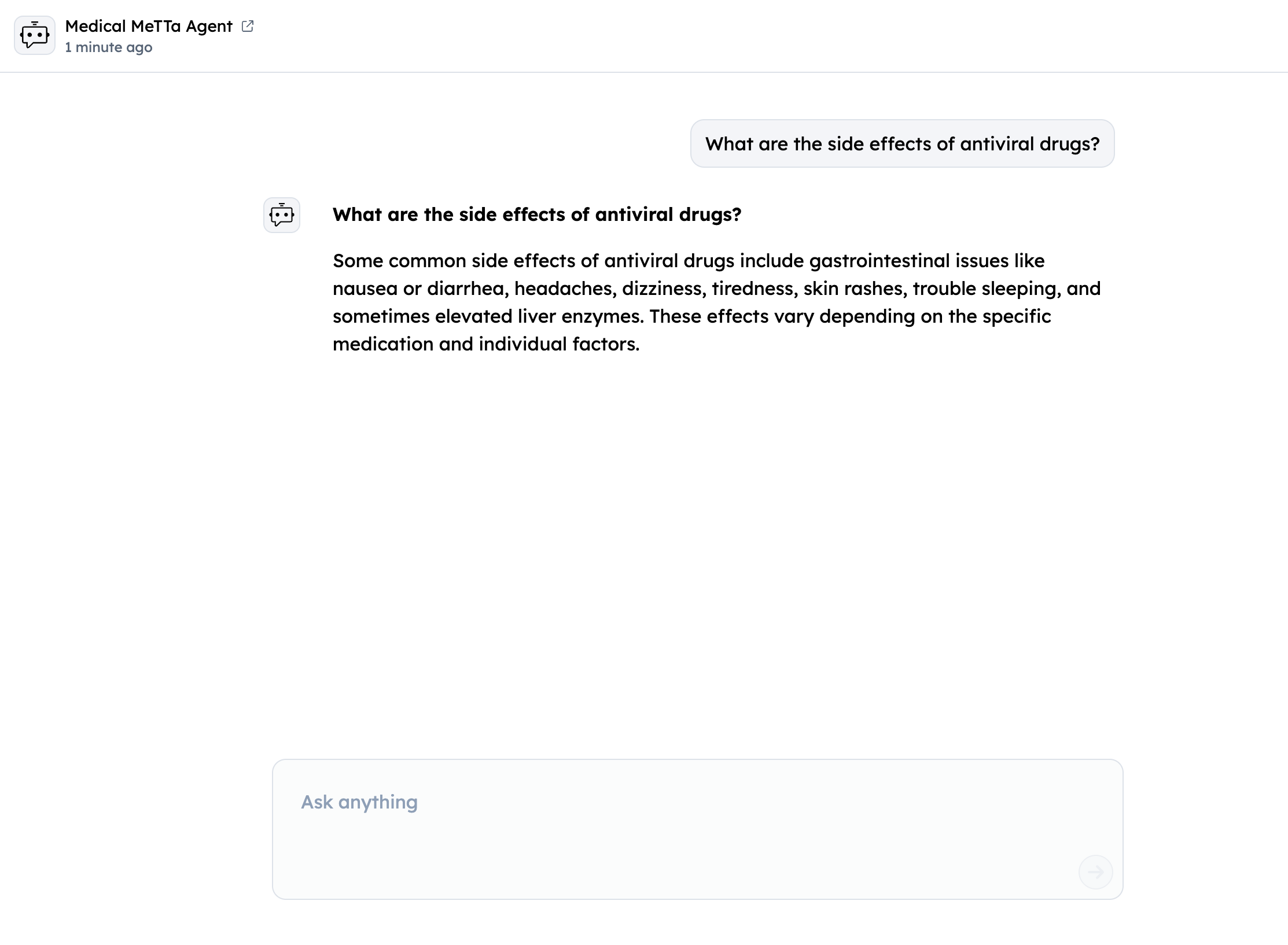
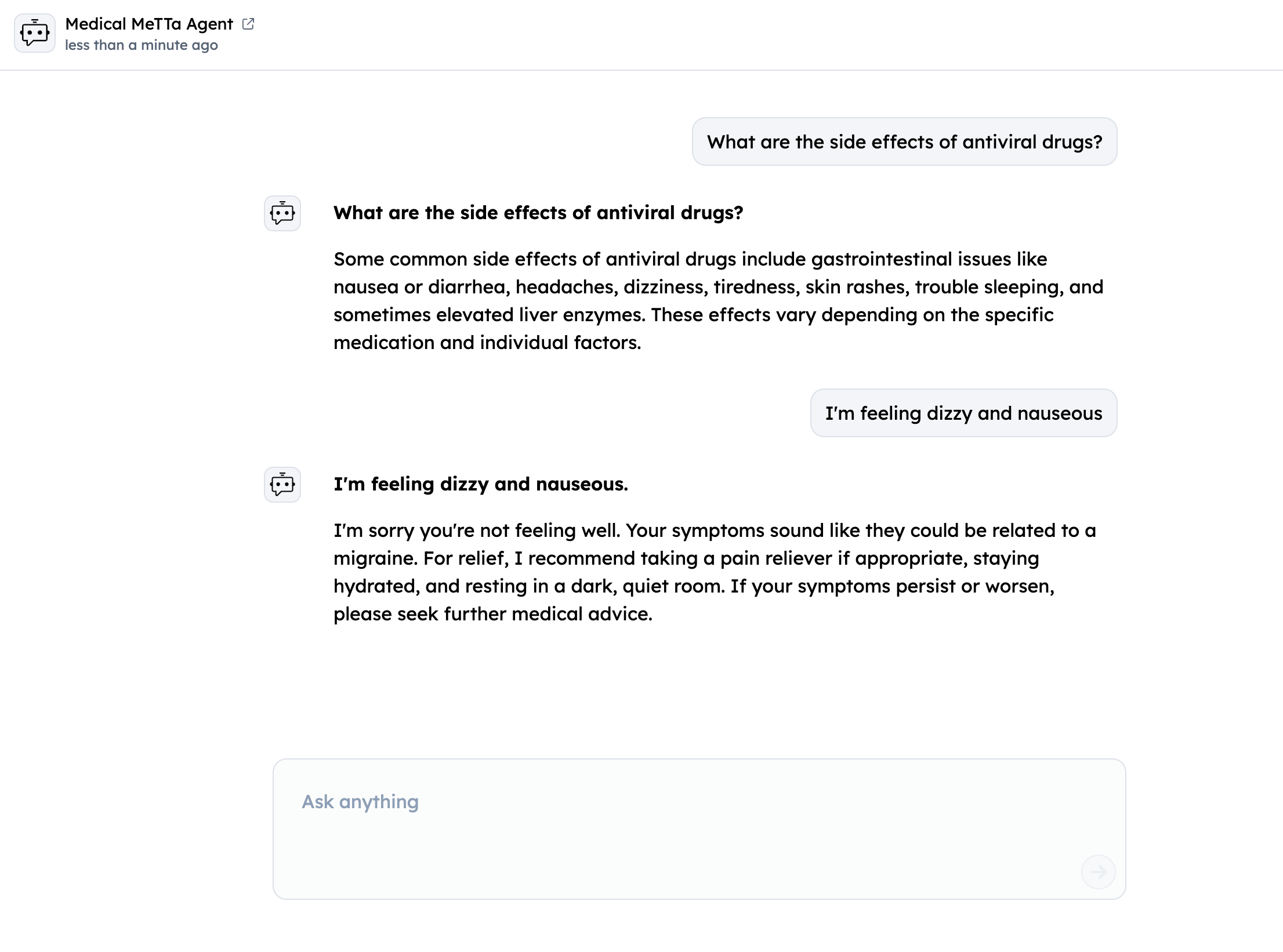
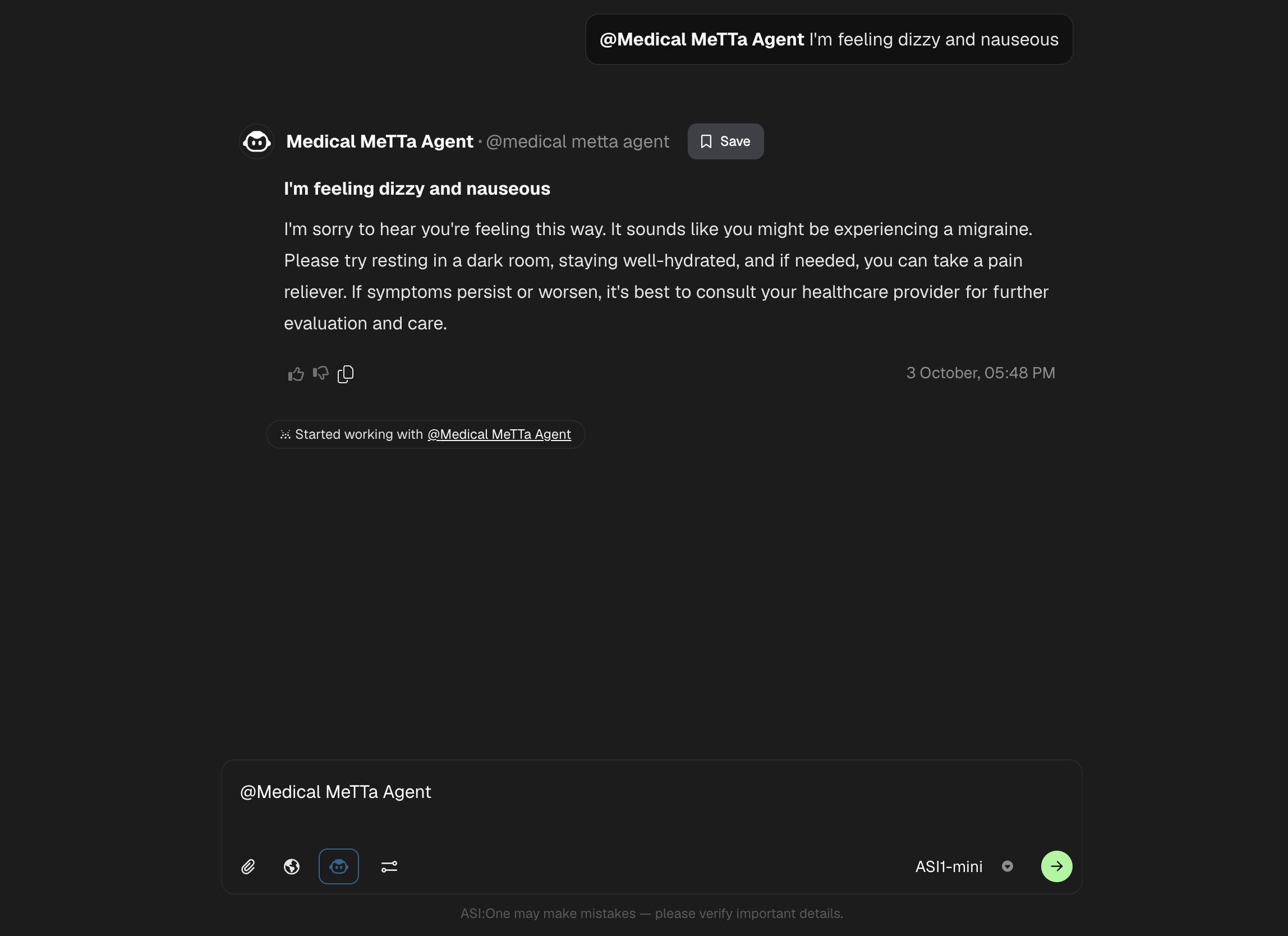
Query your Agent from ASI1 LLM
- Login to the ASI1 LLM, either using your Google Account or the ASI1 Wallet and Start a New Chat.
- Toggle the "Agents" switch to enable ASI1 to connect with Agents on Agentverse.